Executable Science: Research Software Engineering Practices for Replicating Neuroscience Findings
DOI: https://doi.org/10.1145/3736731.3746147
ACM REP '25: ACM Conference on Reproducibility and Replicability, Vancouver, BC, Canada, July 2025
Computational approaches are central to deriving insights in modern science. In this interdisciplinary convergence, researchers are increasingly expected to share research code and data along with published results. However, with software engineering practices still predominantly an overlooked component in research, it remains unclear whether sharing data and code alone is sufficient for reproducibility in practice. We set out to reproduce and replicate evaluation results of a publicly available human neuroimaging dataset recorded when participants listened to short stories. Using the code and data shared with the paper, we predicted the brain activity of the participants with semantic embeddings of words in stories. We successfully reproduced the original results, showing that model performance improved with more training data. However, when we replicated the analysis with our own code, the model's performance was much lower than reported in the paper. We traced this difference to the discrepancy between method description and its implementation in shared code. While access to original code proved crucial for our ability to replicate the results, barriers such as a complex code structure and insufficient documentation hindered our progress. We argue that software engineering practices are essential for reproducible and reusable research. We discuss components of our workflow and specific practices that effectively mitigate barriers to reproducibility.
ACM Reference Format:
Gabriel Kressin Palacios, Zhuoyang Li, and Kristijan Armeni. 2025. Executable Science: Research Software Engineering Practices for Replicating Neuroscience Findings. In ACM Conference on Reproducibility and Replicability (ACM REP '25), July 29--31, 2025, Vancouver, BC, Canada. ACM, New York, NY, USA 14 Pages. https://doi.org/10.1145/3736731.3746147
1 Introduction
Computational methods are an indispensable part of contemporary science. For example, cognitive computational neuroscience is a nascent discipline using computational models to investigate the brain's representations, algorithms, and mechanisms underlying human intelligent behavior in healthy and clinical populations [54, 67]. As computational techniques become central to scientific discovery, the ability of independent research teams to reproduce and replicate published work has been recognized as a standard for evaluating the reliability of scientific findings [23, 30, 69, 76]. Researchers are now strongly encouraged to share open and reproducible research data, code, and documentation alongside written reports [52, 70, 77, 79], a trend also reflected in cognitive computational neuroscience [16, 28, 40, 80, 81, 86]. Despite this progress, achieving computational reproducibility in research remains a significant practical challenge, due to the increasing complexity of both data and analytical methods, as well as ethical and legal constraints [e.g., 74, 15, 52, 34, 44, 105, 6].
While there is growing momentum toward sharing research artifacts such as data and code, making them available does not guarantee reproducibility [63]. To be truly reproducible, research artifacts must also be reusable, readable, and resilient [27]. Reproduction efforts can falter when artifacts are incomplete or contain errors – such as missing data or faulty scripts [e.g., 78, 44, 71] – or when the documentation is sparse or unclear, making it difficult to rerun analyses with the provided data [105]. In contrast, the software engineering community has long developed best practices for creating robust and reusable general-purpose artifacts [e.g., 93, 9, 62, 98]. However, these practices are not always directly applicable to research settings, where the scope of engineering needs varies from standalone analysis scripts to domain-specific research software tools [5, 27]. Given that much of scientific research is conducted within relatively small, independent teams, a central question arises: what software practices and open-source tools are best suited to support reproducible research?
Here, we share our experience and lessons learned from a small-scale reproducibility project in an academic neuroscience lab. We set out to reproduce and replicate the evaluation of predictive models on a publicly available fMRI dataset, which demonstrated that increasing the amount of training data improved average performance in predicting brain activity at the individual participant level [58]. We conducted three complementary experiments: i) a reproduction experiment using the original shared data and code, ii) a replication experiment using the shared data but re-implementing the analysis with our own code, and iii) an extension experiment using the shared data in a novel analysis not reported in the original paper. The main contributions of our work are:
- We successfully reproduced the original results using the shared data and code.
- We identified several challenges when replicating the analysis with our own implementation.
- We describe components of our reproducible workflow and review software engineering practices that can facilitate reproducible research.
2 Methods
In what follows, we provide a high-level overview of the methodology, the data used, and the analysis design for our experiments following the original experiment and dataset [58]. All of our analysis and visualization code was implemented in Python 3.12 and the Python scientific stack (NumPy [45], Pandas [64, 92], SciPy [96], scikit-learn [75], matplotlib [48, 91], seaborn [99], pycortex [37]). The data and code for the replication and reproducibility experiments and the documentation is available at standalone OSF1 and GitHub repositories2, respectively.
2.1 Encoding Models
A central goal in computational neuroscience is to test hypotheses about the computations and representations the brain uses to process information. Encoding models address this goal by quantifying the relationship between features of a stimulus and its corresponding neural responses.
In practice, machine learning methods are often used to extract relevant stimulus features (e.g., semantic, visual, or acoustic), which are then used in statistical models (typically linear regression [50]) to predict neural activity [68]. By comparing the predictive performance of different encoding models (e.g., from different stimulus features across different brain areas), researchers can draw inferences about the spatial and temporal organization of brain function [29, 55]. Encoding models are also foundational for brain-computer interfaces, including clinical applications such as speech prostheses, which aim to reconstruct intended speech from recorded brain activity [88].
Formally, an encoding model estimates a function that maps stimulus features X to brain responses Y. The model typically takes a linear form:
Model estimation involves fitting $\hat{{\bf W}}$ to predict brain activity $\hat{{\bf Y}}_{\text{train}}$ from features Xtrain using statistical methods such as ridge regression. Model performance is then evaluated on held-out test data by comparing predicted brain activity $\hat{{\bf Y}}_{\textsf {test}}$ to observed brain activity Ytest using a scoring function such as Pearson correlation: $r = \textsf {correlation}(\hat{{\bf Y}}_\text{test}, {\bf Y}_\text{test})$.
| Parameter | Lebel et al. [58] | Reproduction | Replication |
|---|---|---|---|
| model | ridge regression | ridge regression | ridge regression |
| model implementation | custom script | custom script | RidgeCV() from scikit-learn |
| cross-validation scheme | leave-one-story-out | leave-one-story-out | leave-one-story-out |
| Ntotal stories | 26 | 26 | 26 |
| α | np.logspace(1, 3, 10) | np.logspace(1, 3, 10) | np.logspace(1, 3, 10) |
| scoring function | Pearson correlation | Pearson correlation | Pearson correlation |
| nchunks | 125 | 10 | n/a |
| chunklen | 40 | 10 | n/a |
| n_delays | 4 | 4 | 4 |
| nboots | 50 | 20 | n/a |
| TRs trimmed | 5 | 5 | 10 |
2.2 Dataset
The publicly available fMRI dataset provided by Lebel et al. [58] recorded blood-oxygen-level-dependent (BOLD) signals while participants listened to 27 natural narrative stories. The dataset is available via the OpenNeuro repository3.
2.2.1 Preprocessed fMRI responses (${\bf Y}_{\textsf {fmri}}$). Because the focus of our work was on encoding models, we chose to use the preprocessed fMRI data provided by Lebel et al. [58]. As a result, our reproducibility efforts did not involve preprocessing of raw fMRI data, which includes steps such as motion correction, cross-run alignment, and temporal filtering. We limited our analysis to a subset of three participants (S01, S02, S03) because they showed the highest signal quality and highest final performance in the original paper. We reasoned that this data would be sufficient to evaluate reproducibility and replicability, as higher quality data would allow us to make more nuanced evaluations in case we found performance discrepancies, and that this data is more likely to be used by other researchers. We accessed the data using the DataLad data management tool [43], as recommended by Lebel et al. [58]. Time series were available as .h5p files for each participant and each story. Following the original report, the first 10 seconds (5 TRs) of each story were trimmed to remove the 10-second silence period before the story began.
Hemodynamic response estimation. The fMRI BOLD responses are thought to represent temporally delayed (on the scale of seconds) and slowly fluctuating components of the underlying local neural activity [59]. Following Lebel et al. [57] we constructed predictor features for timepoint t by concatenating stimulus features from time points t − 1 to t − 4 (ndelay = 4) to account for the delayed neural response. For timepoints where t − k < 0, zero vectors were used as padding. This resulted in a predictor matrix X ∈ RT × 4D. Although this increases computational cost, it enables the regression model to capture the shape of the hemodynamic response function [17] underlying the BOLD signal.
2.3 Predictors
2.3.1 Semantic predictor (${\bf X}_{\textsf {semantic}}$). To model brain activity related to aspects of linguistic meaning understanding during story listening, Lebel et al. [58] used word embeddings — high-dimensional vectors capturing distributional semantic properties of words based on their co-occurrences in large collections of text [24].
Extracting word embeddings and timings. For each word in the story, we extracted its precomputed 985-dimensional embedding vector [49] from the english1000sm.hf5 4 data matrix (a lookup table) provided by Lebel et al. [58] and available in the OpenNeuro repository. Words not present in the vocabulary were assigned a zero vector. For every story, this yielded a $\hat{{\bf X}}_{\textsf {semantic}} \in \mathrm{R}^{N^{words} \times 985}$ matrix of word embeddings for each story where Nwords are all individual words in a story. The onset and offset times (in seconds) of words were extracted from *.TextGrid annotation files5 provided with the original dataset.
Aligning word embeddings with BOLD signal. The fMRI BOLD signal was sampled at regular intervals (repetition time, or TR = 2s). To compute a stimulus matrix $\hat{{\bf X}}_{\textsf {semantic}}$ that matched the sampling rate of the BOLD data, following Lebel et al. [57], we first constructed an array of word times Tword by assigning each word a time half-way between its onset and offset time. This was used to transform the embedding matrix (which was at discrete word times) into a continuous-time representation. This representation is zero at all timepoints except for the middle of each word Tword, where it is equal to the embedding vector of the word. We then convolved this signal with a Lanczos kernel (with parameter a = 3 and fcutoff = 0.25 Hz) to smooth the embeddings over time and mitigate high-frequency noise. Finally, we resampled the signal half-way between the TR times of the fMRI data to create the feature matrix used for regression, ${\bf X}_{\textsf {semantic}} \in \mathrm{R}^{N^{TRs} \times N^{dim}}$.
2.3.2 Sensory predictor (${\bf X}_{\textsf {sensory}}$). To benchmark the performance of our semantic predictor, we developed an additional sensory encoding model based on the audio envelope of the stories (not reported in the original work [58]). The audio envelope was computed by taking the absolute value of the hilbert-transformed wavfile data. For each story, the envelope was trimmed by removing the first and last 10 seconds and downsampled to match the fMRI sampling frequency.
2.4 Ridge regression
In the reproduction experiment, we used the ridge regression implementation provided with the shared code. In our replication experiment, we used the scikit-learn library [75], which is a mature library for machine learning with a large user-base and community support. Specifically, we used the RidgeCV() class which performs a leave-one-out cross-validation to select the best value of the α hyperparameter for each target variable (i.e. brain activity time courses in each voxel) prior to fitting the model. We set the possible hyperparamater values to α ∈ np.logspace(1, 3, 10) as in the original report [58]. The α value that resulted in the highest product-moment correlation coefficient was then used by RidgeCV() as the hyperparameter value to fit the model.
2.5 Evaluation
2.5.1 Cross-validation. Cross-validation was performed at the story level to ensure the independence of training and test data. Specifically, to construct the training set, we randomly sampled without replacement a subset of Ntrain size stories from the Ntotal stories = 26 training set pool and held out one constant story (Where there's smoke) as the test dataset in each fold. This random sampling process was repeated $N^{\textsf {repeat}} = 15$ times, with a new selection of training stories from Ntotal stories in each iteration as described in the original report [58]. To evaluate the effect of training set size on model performance, we varied Ntrain size ∈ {1, 3, 5, 7, 9, 11, 13, 15, 19, 21, 23, 25}.
The predictor features for both the training and test sets were z-scored prior to ridge regression using normalization parameters computed only from the training set to prevent any statistical information leaking from the test data into the training set, maintaining the integrity of our evaluation procedure.
2.5.2 Performance metrics. The performance of the encoding model was quantified as the Pearson correlation between observed and predicted BOLD responses on the held-out test story. To obtain a more reliable estimate, we averaged encoding model performance across 15 repetitions, thereby reducing the risk of performance being biased by any particular split of the data.
3 Results
3.1 Reproduction experiment
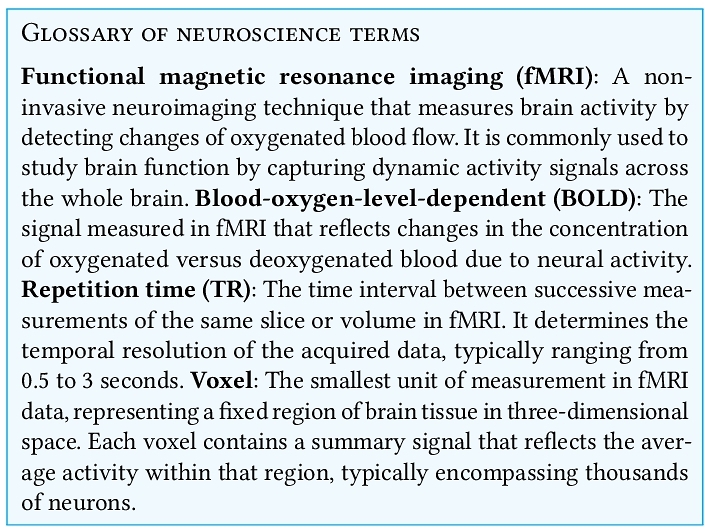
We used the original data and code to reproduce the experimental results. Since the provided code contained the core regression component but lacked the capability to generate figures, we integrated it into our own codebase. We retained the default parameters, except for parameters nboots, chunklen, and nchunk 6 which were set to 20, 10, and 10, respectively (see Table 1).
We successfully reproduced published results. Fig. 1 shows results of our reproduction experiment (panel B) alongside the originally published figures (panel A). Using the shared code and data for the three best performing participants (S01, S02, S03), we reproduced the reported results with participant S03 showing the highest average test-set correlation at r ≈ 0.08, followed by S02 at ≈ 0.07, and S01 at ≈ 0.06.
Model performance increased with larger training dataset sizes. Our reproduction further confirmed that model performance depends on the amount of training data. For the best performing model, average performance nearly tripled (from 0.03 to 0.09) between the smallest (Nstories = 1) and the largest (Nstories = 25) training sets.
Highest performance in the brain areas comprised the language network. Examining the spatial distribution of model performance across individual voxels, we expected the highest performance to occur in regions known to process language. The language network spans multiple lobes, including bilateral temporal, frontal, and parietal cortices [35, 36, 42]. For the best performing model (for participant S02 at Nstories = 25, Fig. 1, panel E), peak performance was achieved for voxels in the left temporal cortex (correlation approximately 0.5), followed by frontal and parietal regions, consistent with the original report.
3.2 Replication experiment
We replicated the spatial and training set size effects, but not the effect size. The results for an example subject are shown in Fig. 1, Panel C. We successfully reproduced the training set size effect and the overall spatial pattern. The best performing voxels were found in bilateral temporal, parietal, and prefrontal cortices, consistent with the original report and our replication. As in the original report, our reproduction results broadly confirmed that the model trained on larger datasets (i.e., more stories) showed better performance on the held-out story. However, our reproduction pipeline yielded substantially lower effect sizes than the original results. Our best performing model achieved only 0.05 average performance compared to 0.09 in the original report and replication. While our models captured the language-related brain activity in relevant regions, they underperformed relative to the original results.
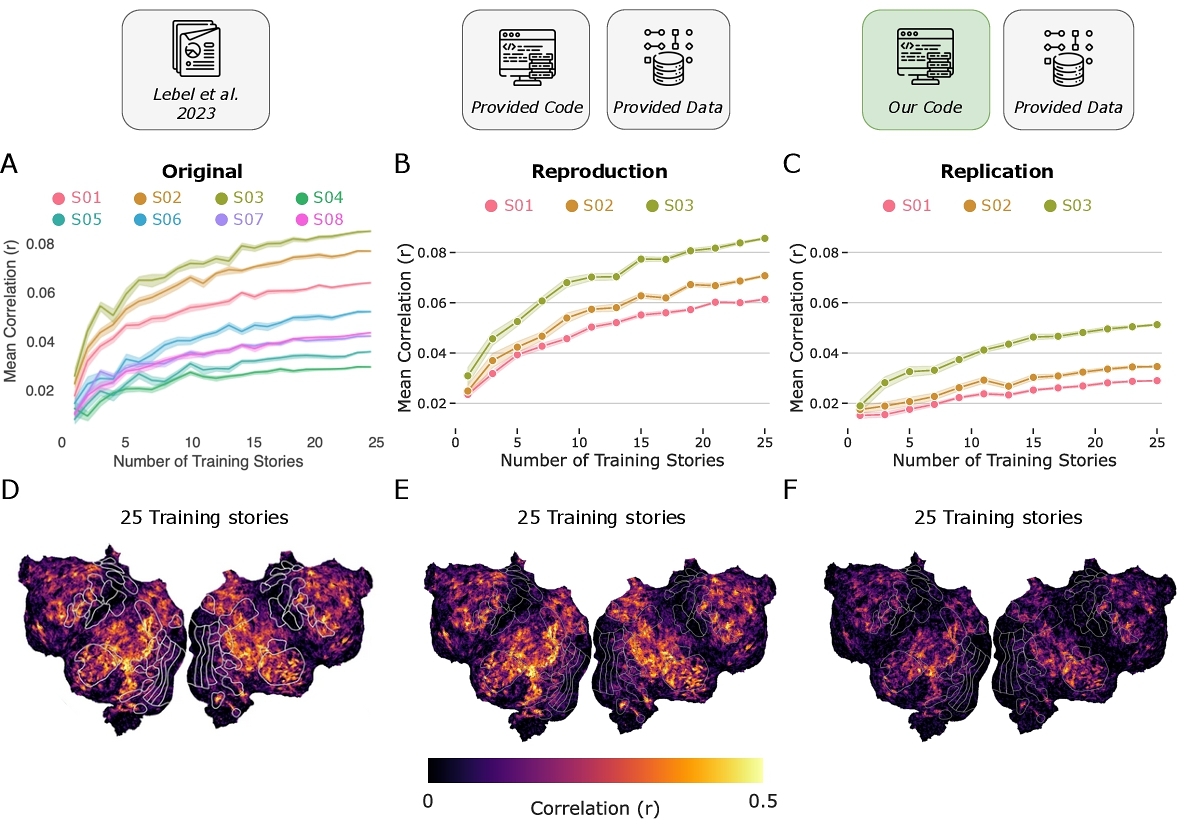
Why did replication results diverge? We identified two key distinctions between the original code and our implementation: 1) instead of standard k-fold cross-validation, the original code used a chunking and bootstrapping strategy [49] for hyperparameter optimization; 2) singular value decomposition (SVD) was applied to remove low-variance components from the regressor matrix prior to regression. To isolate the effects of these differences, we selectively incorporated elements from the original code into our replication pipeline (Fig. 2, panel A) and compared model performance against our replication results (Fig. 2, panel B). We found that incorporating only the chunking and bootstrapping strategy did not improve performance (Fig. 2, panel C). In contrast, fitting ridge regression after SVD — using the function from the original code — restored model performance to the level reported in the original paper (Fig. 2, panel D). These results suggest that the custom ridge regression implementation was essential for reproducing the original results.
3.3 Extension: Sensory encoding model
One of the core motivations behind reproducible computational research is that it should, in principle, allow researchers to extend and build on the shared work. To further benchmark our replication pipeline, we explored the feasibility of performing a small but meaningful extension to the original semantic encoding model.
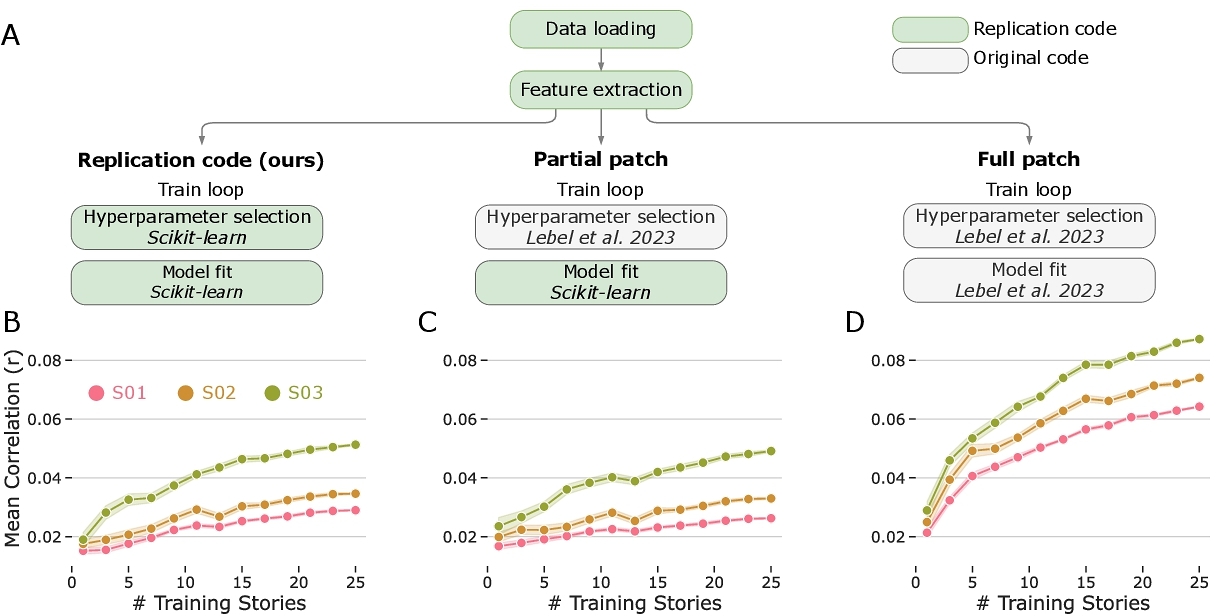
Processing of semantic features in language input is considered a high-level cognitive process that engages a broadly distributed set of brain regions [11, 49]. Next to the semantic interpretation of speech, a narrower set of brain areas near the temples, known as superior temporal cortex, processes speech in terms of its low-level acoustic information (e.g., phonemes, syllables) [47, 73, 82]. To model this, we implemented a simpler encoding model based solely on a single one-dimensional acoustic feature of the stimulus, namely, instantaneous fluctuations of the audio envelope (see Section 2.3.2) which we termed “sensory model”.
In this experiment we did not have prior results on the same dataset to benchmark against. Instead, we constructed a simple control condition by randomly shuffling the target variable (BOLD signals) across time and repeating the regression fitting procedure. The performance of the shuffled regression showed virtually no correlation with fMRI data (“x” markers in Fig. 3) suggesting that sensory models uncovered meaningful neural responses to the audio envelope.
Overall, the results of the sensory model (Fig. 3) are broadly consistent with prior work. Not only did the model perform well above the shuffled baseline, it also captured activity in a restricted set of brain areas, with peak performance localized to the auditory cortex in both hemispheres. Prior work has reliably shown that low-level acoustic features strongly modulate activity localized to auditory cortical regions [47, 73, 82].
4 Discussion
Reproducible computational science ensures scientific integrity and allows researchers to efficiently build upon past work. Here, we report the results of a reproduction and replication project in computational neuroscience, a scientific discipline with computational techniques at its core [54]. We set out to reproduce and replicate evaluation results of a publicly available neuroimaging dataset recorded while participants listened to short stories [58]. Using the code shared with the paper, we reproduced the results showing that predicting brain activity with semantic embeddings of words in stories improved with increasing size of training datasets in each participant. When attempting to replicate the results by implementing the original analysis with our own code, our model performance was much lower than the published results. Accessing the original code, we were able to confirm that the discrepancy was due to the differences in implementation of the regression function.
4.1 Reproduction was largely frictionless, replication and extension were not
There were several examples of good practices that made our reproduction and replications attempts easier. First, Lebel et al. [58], distributed their dataset through a dedicated repository for data sharing [61] and structured the dataset following a domain-specific community standard [81] which made it possible to use a data management software [43] for accessing it. While this might appear a trivial note given that the original work is a dataset descriptor with data sharing as it core objective, it warrants an emphasis as its benefits apply to any empirical work. Specifically, their approach made it easy for us to access the dataset programmatically when building the pipeline (e.g., writing a single script for downloading the data needed in our experiments as opposed to accessing the data interactively). This highlights the importance of data management infrastructure and tools in writing reproducible research code.
Second, the authors provided cursory documentation and instructions on how to use their code for reproducing the results. While the shared code missed some elements that would have been helpful, for example the scripts used for executing the original analyses and the code that produced the figures, the information provided was nevertheless sufficient for us to be able to reproduce their full analysis on the three best participants. In addition, the provided code was modular, with specific analysis steps being implemented as separate routines (for example, the regression module contained the regression fitting functions etc.), making porting specific modules to our code easy and convenient. Analyses in computational neuroscience frequently require custom implementations and workflows, which is common in computational research [5], making a single-analysis-script approach impractical. Navigating the code without basic documentation and sensible organization would have rendered replication much more effortful.
| Reproducibility gain (3R) | Costs | Example tools | ||||||
| Readability | Reuse | Resilience | Technical overhead | Time investment | ||||
|
✓ | ✓ | low | low | Ruff | |||
|
✓ | ✓ | low | mid | mkdocstrings | |||
|
✓ | ✓ | ✓ | low | mid | GitHub, collaborative | ||
|
✓ | ✓ | mid | low | Poetry | |||
|
✓ | mid | mid | git | ||||
|
✓ | ✓ | ✓ | high | high | PyTest | ||
Whereas reproducing the results was mostly achievable, replicating the work with our own code and adapting it for a novel experiment was not without friction. This came to the fore once we started building and evaluating our replication pipeline from the descriptions in the report. For example, when implementing the encoding model, we decided to use a standard machine learning library that conveniently implements ridge regression fitting routines, as it was not apparent from the original paper alone to us which kind of ridge regression was needed. Despite attempting to match the hyperparameter selection procedure, cross-validation scheme, and scoring functions, our encoding models kept underperforming relative to published results. Until we patched our pipeline with the original regression functions, it was unclear just which aspect of our pipeline was causing lower performance. Thus, without the shared code it would have been near impossible for us to troubleshoot our process on the basis of the published report alone.
As another case in point, we encountered challenges when attempting to build on the work and perform a nominally straightforward extension of the original experiment by building an encoding model based on the audio envelope of the stimuli. Whereas the process to temporally align word embeddings and brain data was documented, following the same recipe to align the shared auditory stimuli and brain signal resulted in inconsistencies in the sizes of the to-be-aligned data arrays. We were left to perform our best guess as to the proper alignment by triangulating between stimulus length and number of samples, significantly increasing the effort for an otherwise straightforward extension.
4.2 Tallying software engineering practices for reproducibility in neuroscience
Informed by our reproducibility and replication experience, we argue that software engineering practices, such as code documentation, code review, version control, and code testing, among others, are essential in mitigating barriers to reproducibility beyond what is achievable with data and code sharing alone. In doing so, we rejoin other scientists and software engineers in calling for software engineering maturity in research [e.g., 102, 85, 103, 104, 89, 5, 6, 51]. While the benefits conferred by software engineering practices might appear uncontentious in principle, adoption of any new practice, be it for an individual or in a larger community, is far from straightforward in practice. Absent appropriate incentives, infrastructure, or norms, adopting a new skill or behavior includes a perceived cost (e.g., the time it takes to acquire a new skill) which can be a significant deterrent to adoption [3, 72]. Software engineering practices are no exception: practices will differ in terms of how difficult they are to adopt (i.e., technical overhead required), the time it takes to apply the practice, and the gain they bring for the researcher or the team adopting them.
Table 2 summarizes our own experience in adopting some of the research software practices, how we believe they mitigate the barriers to reproducibility, and their possible costs. In tallying the reproducibility gains, we follow the framework of 3Rs for academic research software proposed by Connolly et al.: research code should be readable, reusable, and resilient [27]. In terms of possible costs, “technical overhead” in Table 2 refers to the degree of novel technical expertise required to adopt a practice. For example, even for someone who regularly writes analysis code, code packaging requires learning the packaging tools, configuration options, the package publishing ecosystem, etc. “Time investment”, on the other hand, refers to the amount of time it takes to subsequently apply a practice once learned7.
Research software specifically can vary in scope on a spectrum between a standalone analysis script and up to a mature software project used by a larger research community [27]. Research software in computational neuroscience typically falls in between the two extremes. Computational neuroscience is a diverse discipline in terms of the scale of investigations (e.g., ranging from single-neuron to whole-brain recordings) and the type of measurements used (e.g., static anatomical images, dynamic multi-channel activity over time, or a combination of different data modalities) [86]. What the analyses have in common, however, is that they are composed of several stages (e.g., preprocessing, model fitting, statistical inference, visualization)[39] such that they frequently require custom routines and workflows [5]. With his context in mind, what aspects of software engineering practices can specifically benefit computational reproducibility in human neuroscience?
Improving readability: Documentation, code formatting, and code review.
Our replication experience showed that methodological details described in the published report were insufficient in detail and our success hinged on our ability to reverse engineer the procedures with the help of shared code. Details matter: given the complexity of analytical pipelines in computational neuroscience, even a nominally small analytical deviation can have large cumulative effects on the results [19, 34, 39, 80]. Insufficient reporting standards in fMRI research have been confirmed in large-scale analyses of published work [19, 41, 80]. Our first set of recommended practices thus centers on code transparency and readability.
Sufficiently documented code significantly boosted our understanding of its purpose and thus our ability to reuse it in replication experiments. Conversely, the lack of such documentation made even the well-structured code difficult to understand and compelled us to perform a line-by-line walkthrough in order to understand the operations on different variables. Given the modular nature of shared code in neuroscience [39], we argue that, apart from inline comments and cursory mentions in papers, all shared code should contain docstrings for major functions and classes and illustrative guides in how they can be invoked. This is particularly necessary for any custom in-house developed code.
Formatting and linting are simple, yet effective methods to improve code readability and to facilitate reusability. Once set up, dedicated code formatters and linters can be used without additional time investment, but deliver substantial benefits: improved code readability, early detection of potentially costly syntax errors, and style consistency. This facilitates collaboration both within and across research teams. The increased readability is not only valuable in collaborative environments, but also in situations were a single researcher reuses to their own code after an extended period. Overall, we recommend that code formatting and linters become a standard practice in research software development.
Building analysis pipelines in neuroscience is seldom straightforward and often requires frequent iteration cycles. This can lead to technical debt and poorly organized code with unused parts which renders it difficult to understand [97]. Code review, a practice of auditing the code for style, quality, and accuracy by peers or developers other than the authors [1, 4], can help mitigating the accrued technical debt. Systematic quality control is a sign of operational maturity in scientific teams dealing with complex analysis projects [51]. Apart from quality control, code review brings other important benefits such as increased author confidence, collegiality, and cross-team learning [84, 95]. We conducted code reviews in the form of brief code walkthroughs after every meeting. Incremental adoption was crucial. Reviewing entire research code can be burdensome, reviewing smaller code chunks, soon after we implemented them, was very doable, improved code clarity, and on multiple occasions allowed us to catch mistakes early. Like documentation, code review is a relatively straightforward practice to adopt insofar that it requires minimal to no technical overhead.
Improving resilience and reuse: Version control, packaging, and testing. We view version control, packaging, and code testing as practices that require greater investment in familiarity with technical tools and possibly steeper learning curves. Version control, a systematic way of recording changes made to files over time [26], is a standard practice in software engineering. While version control does not directly contribute to readability or reusability (in the sense that even extensively versioned code may not be re-executable), it is essential in making the computational research process resilient — in the event of catastrophic changes, current state of files can be reverted to a previous state. We adopted version control in a simple collaborative workflow where we all team members had write access to a joint remote repository.
Reproducible code guarantees that independent researchers can obtain the original results. But that does not imply that the shared code is straightforward to reuse. Reusable code is easy to install and set up, which facilitates adoption [60], and is part of the FAIR principles of research software [22]. Depending on complexity, research code can be made reusable in various ways [26], one option is to distribute it as an installable package: a collection containing the code to be installed, specification of required dependencies, and any software metadata (e.g., author information, project description, etc.). In practice, packaging means organizing your code following an expected directory structure and file naming conventions [100].
In addition to facilitating code reuse by independent researchers, one of the advantages of installable code is that it can be reused within and across your own projects. That turned out to be a crucial design factor in our reproducible workflow (see Section 4.3 below), where we wrapped our figure-making code into separate functions within the package. These functions can be invoked as part of standalone scripts or imported in interactive notebooks and computational reports. The separation between analysis code (package) and its subsequent reuse (in computational notebooks, reports) follows the principle of modular code design, also known as do-not-repeat-yourself (DRY) principle [104]. Most programming languages come with dedicated packaging managers [2]. Whereas Python packaging ecosystem is sometimes perceived as unwieldy8, we found that modern packaging tools, for example Poetry 9 and uv 10, were mostly straightforward to use, did not incur substantial technical overhead, and required moderate time investment (e.g., ∼ 60 mins).
Finally, possibly the most extensive practice we include on our list is code testing. Code testing is a process of writing dedicated routines that test specific parts of code or entire workflows for accuracy and syntactic correctness. Test-driven development in pure software is a mature discipline [8]. It provides the broadest reproducibility benefits, yet testing research code comes with its own specific considerations and challenges [33]. To highlight just two, analysis code in empirical research depends on the data which can contain inconsistencies and exceptions making it challenging to write a single test covering all exceptions. In addition, in neuroscience the datasets tend to be large (on the orders of tens or hundreds of gigabytes) and given the need for tests to be performed frequently, dedicated test datasets would need to be created that appropriately mock real dataset characteristics while remaining lightweight. Second, the research process is iterative (e.g., output of one analysis stage shapes the analysis at the next stage) and the boundaries between the development and deployment phases are frequently blurry [97], leading to challenging testing decisions and frequent need for updating the test suite. Developing testing code demands upfront knowledge of requirements and substantial investment of time and dedicated software expertise, neither of which are currently plentiful in neuroscience.
Testing is the final software engineering practice on our list for a reason; we only started writing limited unit tests and basic package installation tests later in the project phase, once the workflows matured and required less frequent changes. Despite the challenges, we see clear benefits in striving towards test-driven development in research as it forces researchers to not only ascertain accuracy of code, but also to to think carefully about code design, and render explicit code assumptions.
4.3 Towards reproducible scientific publishing workflows
Fig. 4 shows a high-level overview of our adopted code, documentation, and publishing workflow. It shows three main streams: i) packaged code and documentation, ii) computation, storage, and archiving, and iii) computational report. This architecture allows research code (e.g., analysis and plotting scripts) to be developed and versioned in one place and independently of the deployment (HPC) and the publishing streams. The workflow is semi-automatic. For example, documentation and report are deployed and rebuilt automatically upon changes to repositories via GitHub actions and GitHub Pages. However, if analyses change, the researcher must redeploy the jobs on the computing cluster, download the new results, re-execute the report pipeline, and update the remote repositories.
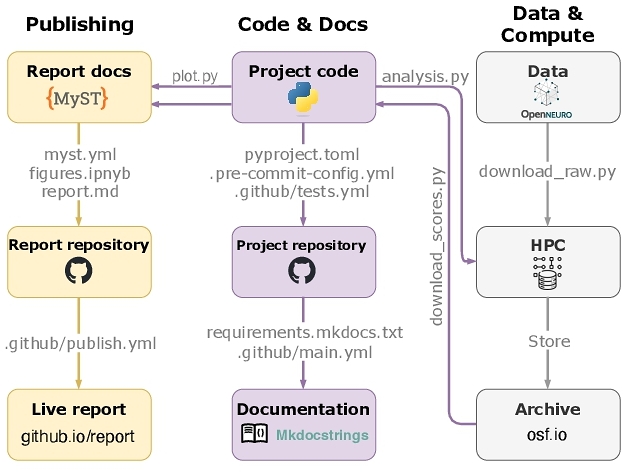
Package and documentation. The analysis package contains the separate .py modules corresponding to distinct analysis stages (e.g., data.py for loading the data, regression.py for model fitting, etc.) which provide the relevant functions (e.g., data.load_fmri, regression.ridge_regression, etc.). The analysis code is locally installable as a python package (via Poetry11) meaning that after downloading, the user can install the code and dependencies, for example using either poetry install or via pip install -e. The code was versioned using git and hosted on GitHub, licensed with permissive MIT License.
We used MkDocs12 to parse the contents or the ./docs folder in the project repository and to render the documentation website. We used MkDocstrings extension that automatically parsed analysis code docstrings and included it as part of the documentation website. The documentation site was published via GitHub Pages and set to redeploy automatically upon updates to the code repository via a GitHub actions.
Data and computation. All our analyses were deployed on a high-performance computing (HPC) cluster. Because the encoding models are fit across the entire brain (resulting in ≈ 103 target variables in a regression model) using high-dimensional predictors (e.g., frequently several hundred dimensions), they require sufficiently powered computing infrastructure. For example, to fit a model on the largest training set size with 25 stories, we requested about 60 gigabytes of memory (RAM). This required fitting multiple models in parallel in a separate deployment stream and precluded, for example, executing all the analyses sequentially in an interactive Jupyter notebook session.
Publishing. A separate repository was used to visualize the results and publish an interactive research report13 using the MyST Markdown framework [25]. MyST allows users to author a computational report in a markdown file, providing all the functionality for technical writing (e.g., citations, cross-references, math rendering, etc.). A report file (report.md) is paired with a Jupyter notebook which imports the plots.py module and reuses the code that creates the figures. Because the notebook cells outputting the figures are labeled (following MyST syntax, see Listing 1), they can be referenced and reused in the report.
4.4 Limitations
End-to-end reproducibility. From the perspective of reproducibility, our workflow falls short in certain respects. Fitting all encoding models reported in this work necessitates access to a high-performance computing (HPC) cluster14, which is not universally available to all research institutions or individual researchers. However, researchers without access to HPC or substantial parallelism can still reproduce the figures from precomputed results (i.e., encoding model performance scores) or reproduce subsets of the full analyses. We expect that with advances of computing resources and infrastructure [e.g., 46] such barriers will become progressively less limiting, but presently remain a barrier.
Other aspects of the software engineering and scientific computing landscape. We focused on what we believe are software practices that directly mitigate barriers to reproducibility in a research lab setting. In doing so, we did not discuss other aspects of reproducible research such as data management [90], workflow management [101], licensing [66], software metadata [22], containerization [2, 65], or cloud-computing [10]. We think these are important and exciting avenues for reproducible research and refer interested readers to cited references for further information about these aspects.
Impacts of AI-assisted software development. A common deterrent in adoption of software engineering practices in research is that these are time-consuming. Recent developments in generative artificial intelligence (AI), computing systems that generate code from prompts given in natural language, are reshaping how software engineers and researchers produce and evaluate code. How will AI-assisted programming affect reproducible research?
At the time of this writing, the landscape is still evolving, confined to individual experimentation and prototyping. In our work, the use of AI-assisted programming was left to individual setup of each author. Our limited experience confirmed that AI-assisted programming reduced friction in specific tasks, such as writing documentation. While there are good reasons to expect that judicious use of AI-assisted programming will facilitate aspects of reproducible work that are currently considered time-consuming [32], using unchecked outputs of an over-confident code generation system can lead to erroneous code [e.g., 56]. Given the broad technical, ethical, and legal challenges when it comes to the use of generative AI in science [e.g., 14, 13, 12, 21, 20, 87], it will require dedicated efforts of professional communities to establish sound practices in AI-assisted development for reproducible research.
4.5 Moving forward
We argue that software engineering practices must become a standard component in research to deliver on the promises of reproducible science. Because the scope of engineering needs in research can vary greatly across disciplines and projects, there is no one-size-fits-all approach in terms of what practices should be adopted and when [27]. Below we briefly outline three ways of moving towards adopting software engineering practices for reproducibility.
Starting in research teams and laboratories. Research groups and laboratories occupy an organizational level which allows swifter implementation of new policies than at the larger organizational levels with typically longer processes (e.g., university departments, schools etc.) and can be tailored to the needs of the group. Research teams could adopt a policy that encourages reproducible science, for example that research code (and other research artifacts) be reviewed by peers before papers are submitted [e.g., 7]. A more comprehensive policy are internal reproducibility audits [69], where research artifacts are validated independently by a peer within the team prior to publication. Regardless of the comprehensiveness level, collaborative code review should be viewed as an opportunity for knowledge exchange (coding strategies, new tools, etc.) among team members and strengthening the engineering expertise of the team as opposed to solely an error finding process [95].
Towards integrated research objects and executable science. To bring reproducible computational research to the fore, the research community should move beyond regarding research code and data as accompanying outputs to static reports. Instead, reports, code, data and other artifacts should be embedded as integrated research objects [31] and considered an equally important and credit-worthy output. All components in integrated research objects would undergo peer review process, which would provide the ability to reproduce key analyses and figures with minimal efforts increasing the confidence in the reliability of the results. An exciting avenue to realize this vision are executable research documents that combine text, data, code and visualization. Although the idea has a venerable history [e.g., 23], authoring executable computational reports became more accessible recently through dedicated technical authoring and publishing frameworks such as Quarto 15 and MyST Markdown that we adopted in current work. While numerous challenges remain, for example how to archive nested computational objects and the costs of computing infrastructure [31], the field of computational neuroscience and neuroimaging offers promising initiatives such as the Neurolibre16 preprint server integrating data, code, and runtime environment [53].
Professionalization of research software engineering in science. Given the central role of programming and computational techniques in science today, research software engineering should be considered as a professional discipline with its own set of standards, curricula, recognition, and scholarly communities. Currently, code development in research does not yet enjoy equal professional standards and support as other scholarly activities and outputs, for example, teaching, scientific writing and citation practices [see 38, 83]. Professionalization is a slowly unfolding, incremental generational process requiring change at multiple levels, such as development training opportunities [6], the possibility of a career path and professional communities [18, 94], and incentive structures [6, 72], among others. As a starting point, team leads should encourage interested researchers to engage with professional communities, such as the US Research Software Engineers association in the United States [94], and open source program offices (OSPOs), if one exists, at their institutions.
5 Conclusion
Trustworthy, reliable, and effective scientific progress requires science to be reproducible. Here we reported on our effort to reproduce and replicate models of brain activity in a published fMRI story listening dataset. Despite the results being reproducible with original code, independent replication on the basis of report alone proved challenging and required substantial effort. We argue that to deliver on the promises of open and reproducible science, software engineering practices are essential in mitigating barriers to computational reproducibility and replicability that remain even if data and code are openly shared.
Acknowledgments
We gratefully acknowledge the support of Christopher J. Honey, and the support of the NIH (R01MH119099 and 2P50MH109429) and the NSF (CAREER Award 2238711) awarded to Christopher J. Honey. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. This work was carried out at the Advanced Research Computing at Hopkins (ARCH) core facility (rockfish.jhu.edu), which is supported by the National Science Foundation (NSF) grant number OAC1920103.
Author contributions
The contributions follow the CRediT framework (https://credit.niso.org/): Gabriel Kressin Palacios: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Zhuoyang Li: Conceptualization, Data curation, Formal analysis, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. Kristijan Armeni: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. All authors have read and agreed to the published version of the manuscript.
References
- A.F. Ackerman, L.S. Buchwald, and F.H. Lewski. 1989. Software inspections: an effective verification process. IEEE Software 6, 3 (May 1989), 31–36. https://doi.org/10.1109/52.28121
- Mohammed Alser, Brendan Lawlor, Richard J. Abdill, Sharon Waymost, Ram Ayyala, Neha Rajkumar, Nathan LaPierre, Jaqueline Brito, André M. Ribeiro-dos Santos, Nour Almadhoun, Varuni Sarwal, Can Firtina, Tomasz Osinski, Eleazar Eskin, Qiyang Hu, Derek Strong, Byoung-Do Kim, Malak S. Abedalthagafi, Onur Mutlu, and Serghei Mangul. 2024. Packaging and containerization of computational methods. Nature Protocols 19, 9 (Sept. 2024), 2529–2539. https://doi.org/10.1038/s41596-024-00986-0
- Kristijan Armeni, Loek Brinkman, Rickard Carlsson, Anita Eerland, Rianne Fijten, Robin Fondberg, Vera E Heininga, Stephan Heunis, Wei Qi Koh, Maurits Masselink, Niall Moran, Andrew Ó Baoill, Alexandra Sarafoglou, Antonio Schettino, Hardy Schwamm, Zsuzsika Sjoerds, Marta Teperek, Olmo R van den Akker, Anna van't Veer, and Raul Zurita-Milla. 2021. Towards wide-scale adoption of open science practices: The role of open science communities. Science and Public Policy 48, 5 (Oct. 2021), 605–611. https://doi.org/10.1093/scipol/scab039
- Alberto Bacchelli and Christian Bird. 2013. Expectations, outcomes, and challenges of modern code review. In Proceedings of the 2013 International Conference on Software Engineering(ICSE ’13). IEEE Press, San Francisco, CA, USA, 712–721.
- Gabriel Balaban, Ivar Grytten, Knut Dagestad Rand, Lonneke Scheffer, and Geir Kjetil Sandve. 2021. Ten simple rules for quick and dirty scientific programming. PLOS Computational Biology 17, 3 (March 2021), e1008549. https://doi.org/10.1371/journal.pcbi.1008549 Publisher: Public Library of Science.
- Lorena Barba. 2024. The path to frictionless reproducibility is still under construction. Harvard Data Science Review 6, 1 (Jan. 2024). https://doi.org/10.1162/99608f92.d73c0559 Publisher: The MIT Press.
- Lorena A. Barba. 2012. Reproducibility PI Manifesto. https://doi.org/10.6084/M9.FIGSHARE.104539.V1 Artwork Size: 0 Bytes Pages: 0 Bytes.
- Kent Beck. 2015. Test-driven development: by example (20. printing ed.). Addison-Wesley, Boston.
- Kent Beck, Mike Beedle, Arie van Bennekum, Alistair Cockburn, Ward Cunningham, Martin Fowler, James Grenning, Jim Highsmith, Andrew Hunt, Ron Jeffries, Jon Kern, Brian Marick, Robert C. Martin, Steve Mellor, Ken Schwaber, Jeff Sutherland, and Dave Thomas. 2001. Manifesto for agile software development. http://www.agilemanifesto.org/
- G. Bruce Berriman, Ewa Deelman, Gideon Juve, Mats Rynge, and Jens-S. Vöckler. 2013. The application of cloud computing to scientific workflows: a study of cost and performance. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences 371, 1983 (Jan. 2013), 20120066. https://doi.org/10.1098/rsta.2012.0066 Publisher: Royal Society.
- Jeffrey R. Binder, Rutvik H. Desai, William W. Graves, and Lisa L. Conant. 2009. Where Is the semantic system? A critical review and meta-analysis of 120 functional neuroimaging studies. Cerebral Cortex 19, 12 (Dec. 2009), 2767–2796. https://doi.org/10.1093/cercor/bhp055
- Marcel Binz, Stephan Alaniz, Adina Roskies, Balazs Aczel, Carl T. Bergstrom, Colin Allen, Daniel Schad, Dirk Wulff, Jevin D. West, Qiong Zhang, Richard M. Shiffrin, Samuel J. Gershman, Vencislav Popov, Emily M. Bender, Marco Marelli, Matthew M. Botvinick, Zeynep Akata, and Eric Schulz. 2025. How should the advancement of large language models affect the practice of science?Proceedings of the National Academy of Sciences 122, 5 (Feb. 2025), e2401227121. https://doi.org/10.1073/pnas.2401227121 Publisher: Proceedings of the National Academy of Sciences.
- Abeba Birhane, Atoosa Kasirzadeh, David Leslie, and Sandra Wachter. 2023. Science in the age of large language models. Nature Reviews Physics 5, 5 (May 2023), 277–280. https://doi.org/10.1038/s42254-023-00581-4 Publisher: Nature Publishing Group.
- Rishi Bommasani, Drew A. Hudson, Ehsan Adeli, Russ Altman, Simran Arora, Sydney von Arx, Michael S. Bernstein, Jeannette Bohg, Antoine Bosselut, Emma Brunskill, Erik Brynjolfsson, Shyamal Buch, Dallas Card, Rodrigo Castellon, Niladri Chatterji, Annie Chen, Kathleen Creel, Jared Quincy Davis, Dora Demszky, Chris Donahue, Moussa Doumbouya, Esin Durmus, Stefano Ermon, John Etchemendy, Kawin Ethayarajh, Li Fei-Fei, Chelsea Finn, Trevor Gale, Lauren Gillespie, Karan Goel, Noah Goodman, Shelby Grossman, Neel Guha, Tatsunori Hashimoto, Peter Henderson, John Hewitt, Daniel E. Ho, Jenny Hong, Kyle Hsu, Jing Huang, Thomas Icard, Saahil Jain, Dan Jurafsky, Pratyusha Kalluri, Siddharth Karamcheti, Geoff Keeling, Fereshte Khani, Omar Khattab, Pang Wei Koh, Mark Krass, Ranjay Krishna, Rohith Kuditipudi, Ananya Kumar, Faisal Ladhak, Mina Lee, Tony Lee, Jure Leskovec, Isabelle Levent, Xiang Lisa Li, Xuechen Li, Tengyu Ma, Ali Malik, Christopher D. Manning, Suvir Mirchandani, Eric Mitchell, Zanele Munyikwa, Suraj Nair, Avanika Narayan, Deepak Narayanan, Ben Newman, Allen Nie, Juan Carlos Niebles, Hamed Nilforoshan, Julian Nyarko, Giray Ogut, Laurel Orr, Isabel Papadimitriou, Joon Sung Park, Chris Piech, Eva Portelance, Christopher Potts, Aditi Raghunathan, Rob Reich, Hongyu Ren, Frieda Rong, Yusuf Roohani, Camilo Ruiz, Jack Ryan, Christopher Ré, Dorsa Sadigh, Shiori Sagawa, Keshav Santhanam, Andy Shih, Krishnan Srinivasan, Alex Tamkin, Rohan Taori, Armin W. Thomas, Florian Tramèr, Rose E. Wang, William Wang, Bohan Wu, Jiajun Wu, Yuhuai Wu, Sang Michael Xie, Michihiro Yasunaga, Jiaxuan You, Matei Zaharia, Michael Zhang, Tianyi Zhang, Xikun Zhang, Yuhui Zhang, Lucia Zheng, Kaitlyn Zhou, and Percy Liang. 2022. On the opportunities and risks of foundation models. https://doi.org/10.48550/arXiv.2108.07258 arXiv:2108.07258 [cs].
- Rotem Botvinik-Nezer, Felix Holzmeister, Colin F. Camerer, Anna Dreber, Juergen Huber, Magnus Johannesson, Michael Kirchler, Roni Iwanir, Jeanette A. Mumford, R. Alison Adcock, Paolo Avesani, Blazej M. Baczkowski, Aahana Bajracharya, Leah Bakst, Sheryl Ball, Marco Barilari, Nadège Bault, Derek Beaton, Julia Beitner, Roland G. Benoit, Ruud M. W. J. Berkers, Jamil P. Bhanji, Bharat B. Biswal, Sebastian Bobadilla-Suarez, Tiago Bortolini, Katherine L. Bottenhorn, Alexander Bowring, Senne Braem, Hayley R. Brooks, Emily G. Brudner, Cristian B. Calderon, Julia A. Camilleri, Jaime J. Castrellon, Luca Cecchetti, Edna C. Cieslik, Zachary J. Cole, Olivier Collignon, Robert W. Cox, William A. Cunningham, Stefan Czoschke, Kamalaker Dadi, Charles P. Davis, Alberto De Luca, Mauricio R. Delgado, Lysia Demetriou, Jeffrey B. Dennison, Xin Di, Erin W. Dickie, Ekaterina Dobryakova, Claire L. Donnat, Juergen Dukart, Niall W. Duncan, Joke Durnez, Amr Eed, Simon B. Eickhoff, Andrew Erhart, Laura Fontanesi, G. Matthew Fricke, Shiguang Fu, Adriana Galván, Remi Gau, Sarah Genon, Tristan Glatard, Enrico Glerean, Jelle J. Goeman, Sergej A. E. Golowin, Carlos González-García, Krzysztof J. Gorgolewski, Cheryl L. Grady, Mikella A. Green, João F. Guassi Moreira, Olivia Guest, Shabnam Hakimi, J. Paul Hamilton, Roeland Hancock, Giacomo Handjaras, Bronson B. Harry, Colin Hawco, Peer Herholz, Gabrielle Herman, Stephan Heunis, Felix Hoffstaedter, Jeremy Hogeveen, Susan Holmes, Chuan-Peng Hu, Scott A. Huettel, Matthew E. Hughes, Vittorio Iacovella, Alexandru D. Iordan, Peder M. Isager, Ayse I. Isik, Andrew Jahn, Matthew R. Johnson, Tom Johnstone, Michael J. E. Joseph, Anthony C. Juliano, Joseph W. Kable, Michalis Kassinopoulos, Cemal Koba, Xiang-Zhen Kong, Timothy R. Koscik, Nuri Erkut Kucukboyaci, Brice A. Kuhl, Sebastian Kupek, Angela R. Laird, Claus Lamm, Robert Langner, Nina Lauharatanahirun, Hongmi Lee, Sangil Lee, Alexander Leemans, Andrea Leo, Elise Lesage, Flora Li, Monica Y. C. Li, Phui Cheng Lim, Evan N. Lintz, Schuyler W. Liphardt, Annabel B. Losecaat Vermeer, Bradley C. Love, Michael L. Mack, Norberto Malpica, Theo Marins, Camille Maumet, Kelsey McDonald, Joseph T. McGuire, Helena Melero, Adriana S. Méndez Leal, Benjamin Meyer, Kristin N. Meyer, Glad Mihai, Georgios D. Mitsis, Jorge Moll, Dylan M. Nielson, Gustav Nilsonne, Michael P. Notter, Emanuele Olivetti, Adrian I. Onicas, Paolo Papale, Kaustubh R. Patil, Jonathan E. Peelle, Alexandre Pérez, Doris Pischedda, Jean-Baptiste Poline, Yanina Prystauka, Shruti Ray, Patricia A. Reuter-Lorenz, Richard C. Reynolds, Emiliano Ricciardi, Jenny R. Rieck, Anais M. Rodriguez-Thompson, Anthony Romyn, Taylor Salo, Gregory R. Samanez-Larkin, Emilio Sanz-Morales, Margaret L. Schlichting, Douglas H. Schultz, Qiang Shen, Margaret A. Sheridan, Jennifer A. Silvers, Kenny Skagerlund, Alec Smith, David V. Smith, Peter Sokol-Hessner, Simon R. Steinkamp, Sarah M. Tashjian, Bertrand Thirion, John N. Thorp, Gustav Tinghög, Loreen Tisdall, Steven H. Tompson, Claudio Toro-Serey, Juan Jesus Torre Tresols, Leonardo Tozzi, Vuong Truong, Luca Turella, Anna E. van ‘t Veer, Tom Verguts, Jean M. Vettel, Sagana Vijayarajah, Khoi Vo, Matthew B. Wall, Wouter D. Weeda, Susanne Weis, David J. White, David Wisniewski, Alba Xifra-Porxas, Emily A. Yearling, Sangsuk Yoon, Rui Yuan, Kenneth S. L. Yuen, Lei Zhang, Xu Zhang, Joshua E. Zosky, Thomas E. Nichols, Russell A. Poldrack, and Tom Schonberg. 2020. Variability in the analysis of a single neuroimaging dataset by many teams. Nature 582, 7810 (June 2020), 84–88. https://doi.org/10.1038/s41586-020-2314-9 Publisher: Nature Publishing Group.
- Rotem Botvinik-Nezer and Tor D. Wager. 2023. Reproducibility in neuroimaging analysis: Challenges and solutions. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging 8, 8 (Aug. 2023), 780–788. https://doi.org/10.1016/j.bpsc.2022.12.006
- Geoffrey M. Boynton, Stephen A. Engel, Gary H. Glover, and David J. Heeger. 1996. Linear systems analysis of functional magnetic resonance imaging in human V1. The Journal of Neuroscience 16, 13 (July 1996), 4207–4221. https://doi.org/10.1523/JNEUROSCI.16-13-04207.1996
- Alys Brett, Michael Croucher, Robert Haines, Simon Hettrick, James Hetherington, Mark Stillwell, and Claire Wyatt. 2017. Research Software Engineers: State of the nation report 2017. Technical Report. Zenodo. https://doi.org/10.5281/zenodo.495360
- Joshua Carp. 2012. The secret lives of experiments: methods reporting in the fMRI literature. NeuroImage 63, 1 (Oct. 2012), 289–300. https://doi.org/10.1016/j.neuroimage.2012.07.004
- Gary Charness, Brian Jabarian, and John A. List. 2025. The next generation of experimental research with LLMs. Nature Human Behaviour (March 2025), 1–3. https://doi.org/10.1038/s41562-025-02137-1 Publisher: Nature Publishing Group.
- Monojit Choudhury, Zohar Elyoseph, Nathanael J. Fast, Desmond C. Ong, Elaine O. Nsoesie, and Ellie Pavlick. 2025. The promise and pitfalls of generative AI. Nature Reviews Psychology 4, 2 (Jan. 2025), 75–80. https://doi.org/10.1038/s44159-024-00402-0
- Neil P. Chue Hong, Daniel S. Katz, Michelle Barker, Anna-Lena Lamprecht, Carlos Martinez, Fotis E. Psomopoulos, Jen Harrow, Leyla Jael Castro, Morane Gruenpeter, Paula Andrea Martinez, Tom Honeyman, Alexander Struck, Allen Lee, Axel Loewe, Ben van Werkhoven, Catherine Jones, Daniel Garijo, Esther Plomp, Francoise Genova, Hugh Shanahan, Joanna Leng, Maggie Hellström, Malin Sandström, Manodeep Sinha, Mateusz Kuzak, Patricia Herterich, Qian Zhang, Sharif Islam, Susanna-Assunta Sansone, Tom Pollard, Udayanto Dwi Atmojo, Alan Williams, Andreas Czerniak, Anna Niehues, Anne Claire Fouilloux, Bala Desinghu, Carole Goble, Céline Richard, Charles Gray, Chris Erdmann, Daniel Nüst, Daniele Tartarini, Elena Ranguelova, Hartwig Anzt, Ilian Todorov, James McNally, Javier Moldon, Jessica Burnett, Julián Garrido-Sánchez, Khalid Belhajjame, Laurents Sesink, Lorraine Hwang, Marcos Roberto Tovani-Palone, Mark D. Wilkinson, Mathieu Servillat, Matthias Liffers, Merc Fox, Nadica Miljković, Nick Lynch, Paula Martinez Lavanchy, Sandra Gesing, Sarah Stevens, Sergio Martinez Cuesta, Silvio Peroni, Stian Soiland-Reyes, Tom Bakker, Tovo Rabemanantsoa, Vanessa Sochat, Yo Yehudi, and RDA FAIR4RS WG. 2022. FAIR Principles for Research Software (FAIR4RS Principles). (May 2022). https://doi.org/10.15497/RDA00068 Publisher: Zenodo.
- Jon F. Claerbout and Martin Karrenbach. 1992. Electronic documents give reproducible research a new meaning. In SEG Technical Program Expanded Abstracts 1992. Society of Exploration Geophysicists, 601–604. https://doi.org/10.1190/1.1822162
- Stephen Clark. 2015. Vector space models of lexical meaning. In Handbook of contemporary semantic theory (2 ed.). Wiley-Blackwell, 493–552.
- Rowan Cockett, Franklin Koch, Steve Purves, Angus Hollands, Yuxi Wang, Dylan Grandmont, Chris Holdgraf, Andrea, Jan-Hendrik Müller, Spencer Lyon, Cristian Le, Jim Madge, wwx, Sugan Reden, Yuanhao Geng, Ryan Lovett, Mikkel Roald-Arbøl, Matt McKay, Matthew Brett, M Bussonnier, Mridul Seth, Nicolas M. Thiéry, Raniere Silva, Sarah Brown, Sinan Bekar, Tavin Cole, Thad Guidry, and Toby Driscoll. 2025. jupyter-book/mystmd: v1.3.24. https://doi.org/10.5281/ZENODO.14805610
- The Turing Way Community. 2022. The Turing Way: A handbook for reproducible, ethical and collaborative research. https://doi.org/10.5281/ZENODO.3233853
- Andrew Connolly, Joseph Hellerstein, Naomi Alterman, David Beck, Rob Fatland, Ed Lazowska, Vani Mandava, and Sarah Stone. 2023. Software engineering practices in academia: Promoting the 3Rs—Readability, Resilience, and Reuse. Harvard Data Science Review 5, 2 (April 2023). https://doi.org/10.1162/99608f92.018bf012 Publisher: The MIT Press.
- Saskia EJ de Vries, Joshua H Siegle, and Christof Koch. 2023. Sharing neurophysiology data from the Allen Brain Observatory. eLife 12 (July 2023), e85550. https://doi.org/10.7554/eLife.85550 Publisher: eLife Sciences Publications, Ltd.
- Adrien Doerig, Rowan P. Sommers, Katja Seeliger, Blake Richards, Jenann Ismael, Grace W. Lindsay, Konrad P. Kording, Talia Konkle, Marcel A. J. Van Gerven, Nikolaus Kriegeskorte, and Tim C. Kietzmann. 2023. The neuroconnectionist research programme. Nature Reviews Neuroscience 24, 7 (July 2023), 431–450. https://doi.org/10.1038/s41583-023-00705-w
- David L. Donoho. 2010. An invitation to reproducible computational research. Biostatistics 11, 3 (July 2010), 385–388. https://doi.org/10.1093/biostatistics/kxq028
- Elizabeth DuPre, Chris Holdgraf, Agah Karakuzu, Loïc Tetrel, Pierre Bellec, Nikola Stikov, and Jean-Baptiste Poline. 2022. Beyond advertising: New infrastructures for publishing integrated research objects. PLOS Computational Biology 18, 1 (Jan. 2022), e1009651. https://doi.org/10.1371/journal.pcbi.1009651 Publisher: Public Library of Science.
- Elizabeth DuPre and Russell Alan Poldrack. 2024. The future of data analysis is now: Integrating generative AI in neuroimaging methods development. Imaging Neuroscience 2 (July 2024), 1–8. https://doi.org/10.1162/imaga00241
- Nasir U. Eisty, Upulee Kanewala, and Jeffrey C. Carver. 2025. Testing research software: An in-depth survey of practices, methods, and tools. https://doi.org/10.48550/arXiv.2501.17739 arXiv:2501.17739 [cs].
- Timothy M Errington, Alexandria Denis, Nicole Perfito, Elizabeth Iorns, and Brian A Nosek. 2021. Challenges for assessing replicability in preclinical cancer biology. eLife 10 (Dec. 2021), e67995. https://doi.org/10.7554/eLife.67995 Publisher: eLife Sciences Publications, Ltd.
- Evelina Fedorenko, Anna A. Ivanova, and Tamar I. Regev. 2024. The language network as a natural kind within the broader landscape of the human brain. Nature Reviews Neuroscience 25, 5 (May 2024), 289–312. https://doi.org/10.1038/s41583-024-00802-4 Publisher: Nature Publishing Group.
- Angela D Friederici and Sarah Me Gierhan. 2013. The language network. Current Opinion in Neurobiology 23, 2 (April 2013), 250–254. https://doi.org/10.1016/j.conb.2012.10.002
- James S. Gao, Alexander G. Huth, Mark D. Lescroart, and Jack L. Gallant. 2015. Pycortex: an interactive surface visualizer for fMRI. Frontiers in Neuroinformatics 9 (Sept. 2015). https://doi.org/10.3389/fninf.2015.00023
- Marc-Oliver Gewaltig and Robert Cannon. 2014. Current practice in software development for computational neuroscience and how to improve it. PLOS Computational Biology 10, 1 (Jan. 2014), e1003376. https://doi.org/10.1371/journal.pcbi.1003376 Publisher: Public Library of Science.
- Rick O. Gilmore, Michele T. Diaz, Brad A. Wyble, and Tal Yarkoni. 2017. Progress toward openness, transparency, and reproducibility in cognitive neuroscience. Annals of the New York Academy of Sciences 1396, 1 (May 2017), 5–18. https://doi.org/10.1111/nyas.13325
- Krzysztof J. Gorgolewski, Tibor Auer, Vince D. Calhoun, R. Cameron Craddock, Samir Das, Eugene P. Duff, Guillaume Flandin, Satrajit S. Ghosh, Tristan Glatard, Yaroslav O. Halchenko, Daniel A. Handwerker, Michael Hanke, David Keator, Xiangrui Li, Zachary Michael, Camille Maumet, B. Nolan Nichols, Thomas E. Nichols, John Pellman, Jean-Baptiste Poline, Ariel Rokem, Gunnar Schaefer, Vanessa Sochat, William Triplett, Jessica A. Turner, Gaël Varoquaux, and Russell A. Poldrack. 2016. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific Data 3, 1 (June 2016), 1–9. https://doi.org/10.1038/sdata.2016.44 Number: 1 Publisher: Nature Publishing Group.
- Qing Guo, Melissa Parlar, Wanda Truong, Geoffrey Hall, Lehana Thabane, Margaret McKinnon, Ron Goeree, and Eleanor Pullenayegum. 2014. The reporting of observational clinical functional magnetic resonance imaging studies: A systematic review. PLOS ONE 9, 4 (April 2014), e94412. https://doi.org/10.1371/journal.pone.0094412 Publisher: Public Library of Science.
- Peter Hagoort. 2019. The neurobiology of language beyond single-word processing. Science 366, 6461 (Oct. 2019), 55–58. https://doi.org/10.1126/science.aax0289
- Yaroslav Halchenko, Kyle Meyer, Benjamin Poldrack, Debanjum Solanky, Adina Wagner, Jason Gors, Dave MacFarlane, Dorian Pustina, Vanessa Sochat, Satrajit Ghosh, Christian Mönch, Christopher Markiewicz, Laura Waite, Ilya Shlyakhter, Alejandro De La Vega, Soichi Hayashi, Christian Häusler, Jean-Baptiste Poline, Tobias Kadelka, Kusti Skytén, Dorota Jarecka, David Kennedy, Ted Strauss, Matt Cieslak, Peter Vavra, Horea-Ioan Ioanas, Robin Schneider, Mika Pflüger, James Haxby, Simon Eickhoff, and Michael Hanke. 2021. DataLad: distributed system for joint management of code, data, and their relationship. Journal of Open Source Software 6, 63 (July 2021), 3262. https://doi.org/10.21105/joss.03262
- Tom E. Hardwicke, Manuel Bohn, Kyle MacDonald, Emily Hembacher, Michèle B. Nuijten, Benjamin N. Peloquin, Benjamin E. deMayo, Bria Long, Erica J. Yoon, and Michael C. Frank. 2021. Analytic reproducibility in articles receiving open data badges at the journal Psychological Science : an observational study. Royal Society Open Science 8, 1 (Jan. 2021), 201494. https://doi.org/10.1098/rsos.201494
- Charles R. Harris, K. Jarrod Millman, Stéfan J. Van Der Walt, Ralf Gommers, Pauli Virtanen, David Cournapeau, Eric Wieser, Julian Taylor, Sebastian Berg, Nathaniel J. Smith, Robert Kern, Matti Picus, Stephan Hoyer, Marten H. Van Kerkwijk, Matthew Brett, Allan Haldane, Jaime Fernández Del Río, Mark Wiebe, Pearu Peterson, Pierre Gérard-Marchant, Kevin Sheppard, Tyler Reddy, Warren Weckesser, Hameer Abbasi, Christoph Gohlke, and Travis E. Oliphant. 2020. Array programming with NumPy. Nature 585, 7825 (Sept. 2020), 357–362. https://doi.org/10.1038/s41586-020-2649-2
- Soichi Hayashi, Bradley A. Caron, Anibal Sólon Heinsfeld, Sophia Vinci-Booher, Brent McPherson, Daniel N. Bullock, Giulia Bertò, Guiomar Niso, Sandra Hanekamp, Daniel Levitas, Kimberly Ray, Anne MacKenzie, Paolo Avesani, Lindsey Kitchell, Josiah K. Leong, Filipi Nascimento-Silva, Serge Koudoro, Hanna Willis, Jasleen K. Jolly, Derek Pisner, Taylor R. Zuidema, Jan W. Kurzawski, Kyriaki Mikellidou, Aurore Bussalb, Maximilien Chaumon, Nathalie George, Christopher Rorden, Conner Victory, Dheeraj Bhatia, Dogu Baran Aydogan, Fang-Cheng F. Yeh, Franco Delogu, Javier Guaje, Jelle Veraart, Jeremy Fischer, Joshua Faskowitz, Ricardo Fabrega, David Hunt, Shawn McKee, Shawn T. Brown, Stephanie Heyman, Vittorio Iacovella, Amanda F. Mejia, Daniele Marinazzo, R. Cameron Craddock, Emanuale Olivetti, Jamie L. Hanson, Eleftherios Garyfallidis, Dan Stanzione, James Carson, Robert Henschel, David Y. Hancock, Craig A. Stewart, David Schnyer, Damian O. Eke, Russell A. Poldrack, Steffen Bollmann, Ashley Stewart, Holly Bridge, Ilaria Sani, Winrich A. Freiwald, Aina Puce, Nicholas L. Port, and Franco Pestilli. 2024. brainlife.io: a decentralized and open-source cloud platform to support neuroscience research. Nature Methods 21, 5 (May 2024), 809–813. https://doi.org/10.1038/s41592-024-02237-2 Publisher: Nature Publishing Group.
- Gregory Hickok and David Poeppel. 2007. The cortical organization of speech processing. Nature Reviews Neuroscience 8, 5 (May 2007), 393–402. https://doi.org/10.1038/nrn2113
- J. D. Hunter. 2007. Matplotlib: A 2D graphics environment. Computing in Science & Engineering 9, 3 (2007), 90–95. https://doi.org/10.1109/MCSE.2007.55 Publisher: IEEE COMPUTER SOC.
- Alexander G. Huth, Wendy A. de Heer, Thomas L. Griffiths, Frédéric E. Theunissen, and Jack L. Gallant. 2016. Natural speech reveals the semantic maps that tile human cerebral cortex. Nature 532, 7600 (April 2016), 453. https://doi.org/10.1038/nature17637
- Anna A Ivanova, Martin Schrimpf, Stefano Anzellotti, Noga Zaslavsky, Evelina Fedorenko, and Leyla Isik. 2022. Beyond linear regression: mapping models in cognitive neuroscience should align with research goals. Neurons, Behavior, Data analysis, and Theory 1 (Aug. 2022). https://doi.org/10.51628/001c.37507
- Erik C. Johnson, Thinh T. Nguyen, Benjamin K. Dichter, Frank Zappulla, Montgomery Kosma, Kabilar Gunalan, Yaroslav O. Halchenko, Shay Q. Neufeld, Kristen Ratan, Nicholas J. Edwards, Susanne Ressl, Sarah R. Heilbronner, Michael Schirner, Petra Ritter, Brock Wester, Satrajit Ghosh, Maryann E. Martone, Franco Pestilli, and Dimitri Yatsenko. 2024. SciOps: Achieving productivity and reliability in data-intensive research. https://doi.org/10.48550/arXiv.2401.00077 arXiv:2401.00077 [q-bio].
- Anita S. Jwa and Russell A. Poldrack. 2022. The spectrum of data sharing policies in neuroimaging data repositories. Human Brain Mapping 43, 8 (2022), 2707–2721. https://doi.org/10.1002/hbm.25803 _eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/hbm.25803.
- Agah Karakuzu, Elizabeth DuPre, Loic Tetrel, Patrick Bermudez, Mathieu Boudreau, Mary Chin, Jean-Baptiste Poline, Samir Das, Lune Bellec, and Nikola Stikov. 2022. NeuroLibre : A preprint server for full-fledged reproducible neuroscience. https://doi.org/10.31219/osf.io/h89js
- Nikolaus Kriegeskorte and Pamela K. Douglas. 2018. Cognitive computational neuroscience. Nature Neuroscience 21, 9 (Sept. 2018), 1148–1160. https://doi.org/10.1038/s41593-018-0210-5
- Nikolaus Kriegeskorte and Pamela K Douglas. 2019. Interpreting encoding and decoding models. Current Opinion in Neurobiology 55 (April 2019), 167–179. https://doi.org/10.1016/j.conb.2019.04.002
- Arjun Krishna, Erick Galinkin, Leon Derczynski, and Jeffrey Martin. 2025. Importing Phantoms: Measuring LLM Package Hallucination Vulnerabilities. https://doi.org/10.48550/arXiv.2501.19012 arXiv:2501.19012 [cs].
- Amanda LeBel, Lauren Wagner, Shailee Jain, Aneesh Adhikari-Desai, Bhavin Gupta, Allyson Morgenthal, Jerry Tang, Lixiang Xu, and Alexander G. Huth. 2023. An fMRI dataset during a passive natural language listening task. https://doi.org/10.18112/OPENNEURO.DS003020.V2.0.0
- Amanda LeBel, Lauren Wagner, Shailee Jain, Aneesh Adhikari-Desai, Bhavin Gupta, Allyson Morgenthal, Jerry Tang, Lixiang Xu, and Alexander G. Huth. 2023. A natural language fMRI dataset for voxelwise encoding models. Scientific Data 10, 1 (Aug. 2023), 555. https://doi.org/10.1038/s41597-023-02437-z Publisher: Nature Publishing Group.
- Nikos K. Logothetis. 2003. The underpinnings of the BOLD functional magnetic resonance imaging signal. The Journal of Neuroscience 23, 10 (May 2003), 3963–3971. https://doi.org/10.1523/JNEUROSCI.23-10-03963.2003
- Eric J. Ma. 2024. The human dimension to clean, distributable, and documented data science code. Eric J. Ma's Blog (Oct. 2024). https://ericmjl.github.io/blog/2024/10/25/the-human-dimension-to-clean-distributable-and-documented-data-science-code
- Christopher J Markiewicz, Krzysztof J Gorgolewski, Franklin Feingold, Ross Blair, Yaroslav O Halchenko, Eric Miller, Nell Hardcastle, Joe Wexler, Oscar Esteban, Mathias Goncavles, Anita Jwa, and Russell Poldrack. 2021. The OpenNeuro resource for sharing of neuroscience data. eLife 10 (Oct. 2021), e71774. https://doi.org/10.7554/eLife.71774
- Robert C. Martin. 2012. Clean code: a handbook of agile software craftsmanship (repr. ed.). Prentice Hall, Upper Saddle River, NJ Munich.
- Erin C. McKiernan, Lorena Barba, Philip E. Bourne, Caitlin Carter, Zach Chandler, Sayeed Choudhury, Stephen Jacobs, Daniel S. Katz, Stefanie Lieggi, Beth Plale, and Greg Tananbaum. 2023. Policy recommendations to ensure that research software is openly accessible and reusable. PLOS Biology 21, 7 (July 2023), e3002204. https://doi.org/10.1371/journal.pbio.3002204
- Wes McKinney. 2010. Data structures for statistical computing in Python. In Proceedings of the 9th Python in Science Conference, Stéfan van der Walt and Jarrod Millman (Eds.). 56 – 61. https://doi.org/10.25080/Majora-92bf1922-00a
- David Moreau, Kristina Wiebels, and Carl Boettiger. 2023. Containers for computational reproducibility. Nature Reviews Methods Primers 3, 1 (July 2023), 1–16. https://doi.org/10.1038/s43586-023-00236-9 Publisher: Nature Publishing Group.
- Andrew Morin, Jennifer Urban, and Piotr Sliz. 2012. A quick guide to software licensing for the scientist-programmer. PLOS Computational Biology 8, 7 (July 2012), e1002598. https://doi.org/10.1371/journal.pcbi.1002598 Publisher: Public Library of Science.
- Thomas Naselaris, Danielle S. Bassett, Alyson K. Fletcher, Konrad Kording, Nikolaus Kriegeskorte, Hendrikje Nienborg, Russell A. Poldrack, Daphna Shohamy, and Kendrick Kay. 2018. Cognitive computational neuroscience: A new conference for an emerging discipline. Trends in Cognitive Sciences (Feb. 2018). https://doi.org/10.1016/j.tics.2018.02.008
- Thomas Naselaris, Kendrick N. Kay, Shinji Nishimoto, and Jack L. Gallant. 2011. Encoding and decoding in fMRI. NeuroImage 56, 2 (May 2011), 400–410. https://doi.org/10.1016/j.neuroimage.2010.07.073
- National Academies of Sciences, Engineering, and Medicine. 2019. Reproducibility and replicability in science. National Academies Press, Washington, D.C.https://doi.org/10.17226/25303
- Alondra Nelson. 2022. Ensuring free, immediate, and equitable access to federally funded research. https://bidenwhitehouse.archives.gov/wp-content/uploads/2022/08/08-2022-OSTP-Public-Access-Memo.pdf
- Tien Nguyen, Waris Gill, and Muhammad Ali Gulzar. 2025. Are the majority of public computational notebooks pathologically non-executable?https://doi.org/10.48550/arXiv.2502.04184 arXiv:2502.04184 [cs].
- Brian Nosek. 2019. Strategy for culture change. https://www.cos.io/blog/strategy-for-culture-change
- Jonas Obleser and Frank Eisner. 2009. Pre-lexical abstraction of speech in the auditory cortex. Trends in Cognitive Sciences 13, 1 (Jan. 2009), 14–19. https://doi.org/10.1016/j.tics.2008.09.005 Publisher: Elsevier.
- Open Science Collaboration. 2015. Estimating the reproducibility of psychological science. Science 349, 6251 (Aug. 2015), aac4716. https://doi.org/10.1126/science.aac4716
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot, and E. Duchesnay. 2011. Scikit-learn: Machine learning in python. Journal of Machine Learning Research 12 (2011), 2825–2830.
- Roger D. Peng. 2009. Reproducible research and biostatistics. Biostatistics 10, 3 (July 2009), 405–408. https://doi.org/10.1093/biostatistics/kxp014
- Roger D. Peng. 2011. Reproducible research in computational science. Science 334, 6060 (Dec. 2011), 1226–1227. https://doi.org/10.1126/science.1213847 Publisher: American Association for the Advancement of Science.
- João Felipe Pimentel, Leonardo Murta, Vanessa Braganholo, and Juliana Freire. 2019. A large-scale study about quality and reproducibility of jupyter notebooks. In 2019 IEEE/ACM 16th International Conference on Mining Software Repositories (MSR). 507–517. https://doi.org/10.1109/MSR.2019.00077 ISSN: 2574-3864.
- Joelle Pineau, Philippe Vincent-Lamarre, Koustuv Sinha, Vincent Lariviere, Alina Beygelzimer, Florence d'Alche Buc, Emily Fox, and Hugo Larochelle. 2021. Improving reproducibility in machine learning research. Journal of Machine Learning Research 22, 164 (2021), 1–20. http://jmlr.org/papers/v22/20-303.html
- Russell A. Poldrack, Chris I. Baker, Joke Durnez, Krzysztof J. Gorgolewski, Paul M. Matthews, Marcus R. Munafò, Thomas E. Nichols, Jean-Baptiste Poline, Edward Vul, and Tal Yarkoni. 2017. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nature Reviews Neuroscience 18, 2 (Feb. 2017), 115–126. https://doi.org/10.1038/nrn.2016.167 Publisher: Nature Publishing Group.
- Russell A Poldrack and Krzysztof J Gorgolewski. 2014. Making big data open: data sharing in neuroimaging. Nature Neuroscience 17, 11 (Nov. 2014), 1510–1517. https://doi.org/10.1038/nn.3818
- Cathy J. Price. 2010. The anatomy of language: a review of 100 fMRI studies published in 2009. Annals of the New York Academy of Sciences 1191, 1 (March 2010), 62–88. https://doi.org/10.1111/j.1749-6632.2010.05444.x
- Richard McElreath. 2020. Science as amateur software development. https://www.youtube.com/watch?v=zwRdO9_GGhY
- Ariel Rokem. 2024. Ten simple rules for scientific code review. PLOS Computational Biology 20, 9 (Sept. 2024), e1012375. https://doi.org/10.1371/journal.pcbi.1012375 Publisher: Public Library of Science.
- Geir Kjetil Sandve, Anton Nekrutenko, James Taylor, and Eivind Hovig. 2013. Ten simple rules for reproducible computational research. PLoS Computational Biology 9, 10 (Oct. 2013), e1003285. https://doi.org/10.1371/journal.pcbi.1003285
- Terrence J. Sejnowski, Patricia S. Churchland, and J. Anthony Movshon. 2014. Putting big data to good use in neuroscience. Nature Neuroscience 17, 11 (Nov. 2014), 1440–1441. https://doi.org/10.1038/nn.3839 Publisher: Nature Publishing Group.
- Ilia Shumailov, Zakhar Shumaylov, Yiren Zhao, Nicolas Papernot, Ross Anderson, and Yarin Gal. 2024. AI models collapse when trained on recursively generated data. Nature 631, 8022 (July 2024), 755–759. https://doi.org/10.1038/s41586-024-07566-y Publisher: Nature Publishing Group.
- Alexander B. Silva, Kaylo T. Littlejohn, Jessie R. Liu, David A. Moses, and Edward F. Chang. 2024. The speech neuroprosthesis. Nature Reviews Neuroscience 25, 7 (July 2024), 473–492. https://doi.org/10.1038/s41583-024-00819-9 Publisher: Nature Publishing Group.
- Tim Storer. 2017. Bridging the chasm: A survey of software engineering practice in scientific programming. ACM Comput. Surv. 50, 4 (Aug. 2017), 47:1–47:32. https://doi.org/10.1145/3084225
- Carol Tenopir, Natalie M. Rice, Suzie Allard, Lynn Baird, Josh Borycz, Lisa Christian, Bruce Grant, Robert Olendorf, and Robert J. Sandusky. 2020. Data sharing, management, use, and reuse: Practices and perceptions of scientists worldwide. PLOS ONE 15, 3 (March 2020), e0229003. https://doi.org/10.1371/journal.pone.0229003 Publisher: Public Library of Science.
- The Matplotlib Development Team. 2024. Matplotlib: Visualization with Python. https://doi.org/10.5281/ZENODO.13308876
- The Pandas Development Team. 2024. pandas-dev/pandas: Pandas. https://doi.org/10.5281/zenodo.10957263
- David Thomas and Andrew Hunt. 2020. The pragmatic programmer: your journey to mastery (second edition, 20th anniversary edtion ed.). Addison-Wesley, Boston.
- US Research Software Engineer Association and IEEE Computer Society. 2023. Research software engineers: Creating a career path—and a career. https://doi.org/10.5281/ZENODO.10073232
- Anusha M Vable, Scott F Diehl, and M Maria Glymour. 2021. Code review as a simple trick to enhance reproducibility, accelerate learning, and improve the quality of your team's research. American Journal of Epidemiology 190, 10 (Oct. 2021), 2172–2177. https://doi.org/10.1093/aje/kwab092
- Pauli Virtanen, Ralf Gommers, Travis E. Oliphant, Matt Haberland, Tyler Reddy, David Cournapeau, Evgeni Burovski, Pearu Peterson, Warren Weckesser, Jonathan Bright, Stéfan J. van der Walt, Matthew Brett, Joshua Wilson, K. Jarrod Millman, Nikolay Mayorov, Andrew R. J. Nelson, Eric Jones, Robert Kern, Eric Larson, C J Carey, İlhan Polat, Yu Feng, Eric W. Moore, Jake VanderPlas, Denis Laxalde, Josef Perktold, Robert Cimrman, Ian Henriksen, E. A. Quintero, Charles R. Harris, Anne M. Archibald, Antônio H. Ribeiro, Fabian Pedregosa, Paul van Mulbregt, and SciPy 1.0 Contributors. 2020. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nature Methods 17 (2020), 261–272. https://doi.org/10.1038/s41592-019-0686-2
- Marijn van Vliet. 2020. Seven quick tips for analysis scripts in neuroimaging. PLOS Computational Biology 16, 3 (March 2020), e1007358. https://doi.org/10.1371/journal.pcbi.1007358 Publisher: Public Library of Science.
- Hironori Washizaki. 2024. Guide to the Software engineering body of knowledge (SWEBOK Guide) (4.0 ed.). IEEE Computer Society. https://www.computer.org/education/bodies-of-knowledge/software-engineering/
- Michael L. Waskom. 2021. seaborn: statistical data visualization. Journal of Open Source Software 6, 60 (2021), 3021. https://doi.org/10.21105/joss.03021 Publisher: The Open Journal.
- Leah Wasser, David Nicholson, Ariane Sasso, Alexandre Batisse, Simon Molinsky, Chiara Marmo, Filipe Fernandes, Jonny Saunders, Carol Willing, Trevor James Smith, Jesse Mostipak, Jeremy Paige, Nick Murphy, Billy Broderick, Isabel Zimmerman, Moritz E. Beber, and Erik Welch. 2024. pyOpenSci/python-package-guide: pyOpenSci packaging guide v0.4. Zenodo. https://doi.org/10.5281/zenodo.13154979 Version Number: v0.4.
- Sean R. Wilkinson, Meznah Aloqalaa, Khalid Belhajjame, Michael R. Crusoe, Bruno de Paula Kinoshita, Luiz Gadelha, Daniel Garijo, Ove Johan Ragnar Gustafsson, Nick Juty, Sehrish Kanwal, Farah Zaib Khan, Johannes Köster, Karsten Peters-von Gehlen, Line Pouchard, Randy K. Rannow, Stian Soiland-Reyes, Nicola Soranzo, Shoaib Sufi, Ziheng Sun, Baiba Vilne, Merridee A. Wouters, Denis Yuen, and Carole Goble. 2025. Applying the FAIR Principles to computational workflows. Scientific Data 12, 1 (Feb. 2025), 328. https://doi.org/10.1038/s41597-025-04451-9 Publisher: Nature Publishing Group.
- Gregory Wilson. 2006. Where's the real bottleneck in scientific computing?American Scientist 94, 1 (2006), 5. https://doi.org/10.1511/2006.57.3473
- Greg Wilson, D. A. Aruliah, C. Titus Brown, Neil P. Chue Hong, Matt Davis, Richard T. Guy, Steven H. D. Haddock, Kathryn D. Huff, Ian M. Mitchell, Mark D. Plumbley, Ben Waugh, Ethan P. White, and Paul Wilson. 2014. Best practices for scientific computing. PLOS Biology 12, 1 (Jan. 2014), e1001745. https://doi.org/10.1371/journal.pbio.1001745 Publisher: Public Library of Science.
- Greg Wilson, Jennifer Bryan, Karen Cranston, Justin Kitzes, Lex Nederbragt, and Tracy K. Teal. 2017. Good enough practices in scientific computing. PLOS Computational Biology 13, 6 (June 2017), e1005510. https://doi.org/10.1371/journal.pcbi.1005510 Publisher: Public Library of Science.
- Xin Xiong and Ivor Cribben. 2023. The state of play of reproducibility in statistics: An empirical analysis. The American Statistician 77, 2 (April 2023), 115–126. https://doi.org/10.1080/00031305.2022.2131625
Footnote
⁎Both authors contributed equally to this research.
2 https://github.com/GabrielKP/enc
4 https://github.com/OpenNeuroDatasets/ds003020/blob/main/derivative/english1000sm.hf5
5These are structured.txt files that are used with the Praat software for acoustic analysis (https://www.fon.hum.uva.nl/praat/)
6For an explanation of the parameters see the “Code” section of the original paper [57].
7While these two aspects are certainly positively correlated, a practice can require little technical overhead, while still representing non-negligible time investment (e.g., writing documentation) or vice-versa (e.g., code packaging, which is set up only once, but requires dedicated technical know-how that has to be learned).
13 https://kristijanarmeni.github.io/encoders_report/
14While a desktop computer with 64GB of RAM and 16 cores (AMD Ryzen 9 5950X) was sufficient to fit the most resource-demanding encoding model, the process was time-consuming: fitting 15 repetitions of model fitting on one dataset size (out of total 13) for one participant required 5 hours.

This work is licensed under a Creative Commons Attribution 4.0 International License.
ACM REP '25, Vancouver, BC, Canada
© 2025 Copyright held by the owner/author(s).
ACM ISBN 979-8-4007-1958-5/25/07.
DOI: https://doi.org/10.1145/3736731.3746147